Real-time surveillance of viral genomes enables the detection of new variants, the assessment of their impact on infectivity and disease severity, the estimation of rates of spread and routes of transmission using new phylogenetic tools, and evidence-based decision making by public health authorities, but it requires access to timely genomic information. Up-to-date genomic information also facilitates rapid, collaborative and interdisciplinary research and evidence-based public health responses (for example, vaccine development and deployment). Viral genetic changes have enabled transmission of the highly pathogenic avian influenza H5N1 to hundreds of species of birds and mammals1,2,3, leading to repeated animal-to-human transmission events, including 72 reported to the World Health Organization in 2024 (ref. 4). Two recent reports describe patients with severe respiratory infections in Canada5 and the United States6 with viruses that were polymorphic for genetic changes previously predicted by deep mutational scanning to improve binding to human cells7. While these changes may have facilitated within-host viral replication, efficient human-to-human transmission of H5N1 has yet to be observed. However, this may change at any time. Real-time reporting of current H5N1 genomes is crucial, yet we find extensive delays of 7.5 months between H5N1 sample collection and submission to the Global Initiative on Sharing All Influenza Data (GISAID) repository for virus data and associated metadata8,9.
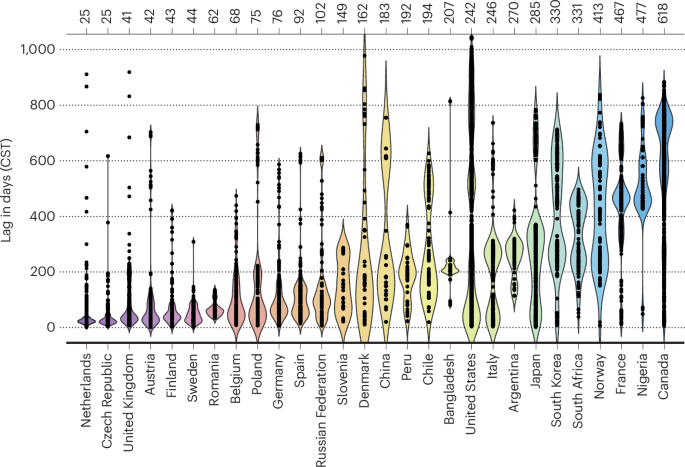
Early on in the COVID-19 pandemic, a previous study10 highlighted lengthy delays in SARS-CoV-2 sequence submissions to GISAID, with an average of 48 days between sample collection and submission. This global analysis highlighted countries with rapid data sharing practices and pointed out others that lagged behind. Many countries subsequently improved pipelines for data submission, with collection-to-submission times (CST) now down to 30 days for samples submitted to GISAID in 2024. As an example, Canada had a CST of 88 days early in the pandemic10, but now has a median CST for SARS-CoV-2 sequences of only 16 days. Such dramatic improvements aided global efforts to track variants and to monitor the spread and public health impacts of COVID-19 over the last few years.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Data availability
The findings of this study are based on metadata accessed from GISAID on 26 February 2025 and includes H5N1 sequences up to 31 December 2024 (list of EPI_SET IDs at https://doi.org/10.55876/gis8.250226wp), H1N1 sequences up to 31 December 2013 (EPI_SET IDs at https://doi.org/10.55876/gis8.250226ad) and H1N1 sequences sampled in 2024 (EPI_SET IDs at https://doi.org/10.55876/gis8.250226zt). Summary data for SARS-CoV-2 were obtained from GISAID’s Dates + Location summary file, limited to submissions in 2024 (EPI_SET IDs at https://doi.org/10.55876/gis8.250226hm).
References
-
Youk, S. et al. Virology 587, 109860 (2023).
Article
CAS
PubMedGoogle Scholar
-
Peacock, T. P. et al. Nature 637, 304–313 (2025).
Article
CAS
PubMedGoogle Scholar
-
Animal and Plant Health Inspection Service. Detections of highly pathogenic avian influenza. https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influenza/hpai-detections (20 March 2025).
-
World Health Organization. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003-2024. https://www.who.int/publications/m/item/cumulative-number-of-confirmed-human-cases-for-avian-influenza-a(h5n1)-reported-to-who–2003-2024–20-december-2024 (12 December 2024).
-
Jassem, A. N. et al. N. Engl. J. Med. 392, 927–929 (2025).
Article
PubMedGoogle Scholar
-
Centers for Disease Control and Prevention. Genetic sequences of highly pathogenic avian influenza A(H5N1) viruses identified in a person in Louisiana. Avian Influenza (Bird Flu) https://www.cdc.gov/bird-flu/spotlights/h5n1-response-12232024.html (2024).
-
Dadonaite, B. et al. PLoS Biol. 22, e3002916 (2024).
Article
CAS
PubMed
PubMed CentralGoogle Scholar
-
Shu, Y. & McCauley, J. Euro Surveill. 22, 30494 (2017).
-
Elbe, S. & Buckland-Merrett, G. Glob. Chall. 1, 33–46 (2017).
Article
PubMed
PubMed CentralGoogle Scholar
-
Kalia, K., Saberwal, G. & Sharma, G. Nat. Biotechnol. 39, 1058–1060 (2021).
Article
CAS
PubMedGoogle Scholar
Download references
Acknowledgements
We gratefully acknowledge all data contributors, that is, the authors and their originating laboratories responsible for obtaining the specimens, and their submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this study is based. We would like to thank Florence Débarre for comments on an earlier draft. We acknowledge the support of the Natural Sciences and Engineering Research Council of Canada (NSERC RGPIN-2022-03726).
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Otto, S.P., Edgerton, S.V. Lengthy delays in H5N1 genome submissions to GISAID.
Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02636-6
Download citation
-
Published:
-
DOI: https://doi.org/10.1038/s41587-025-02636-6